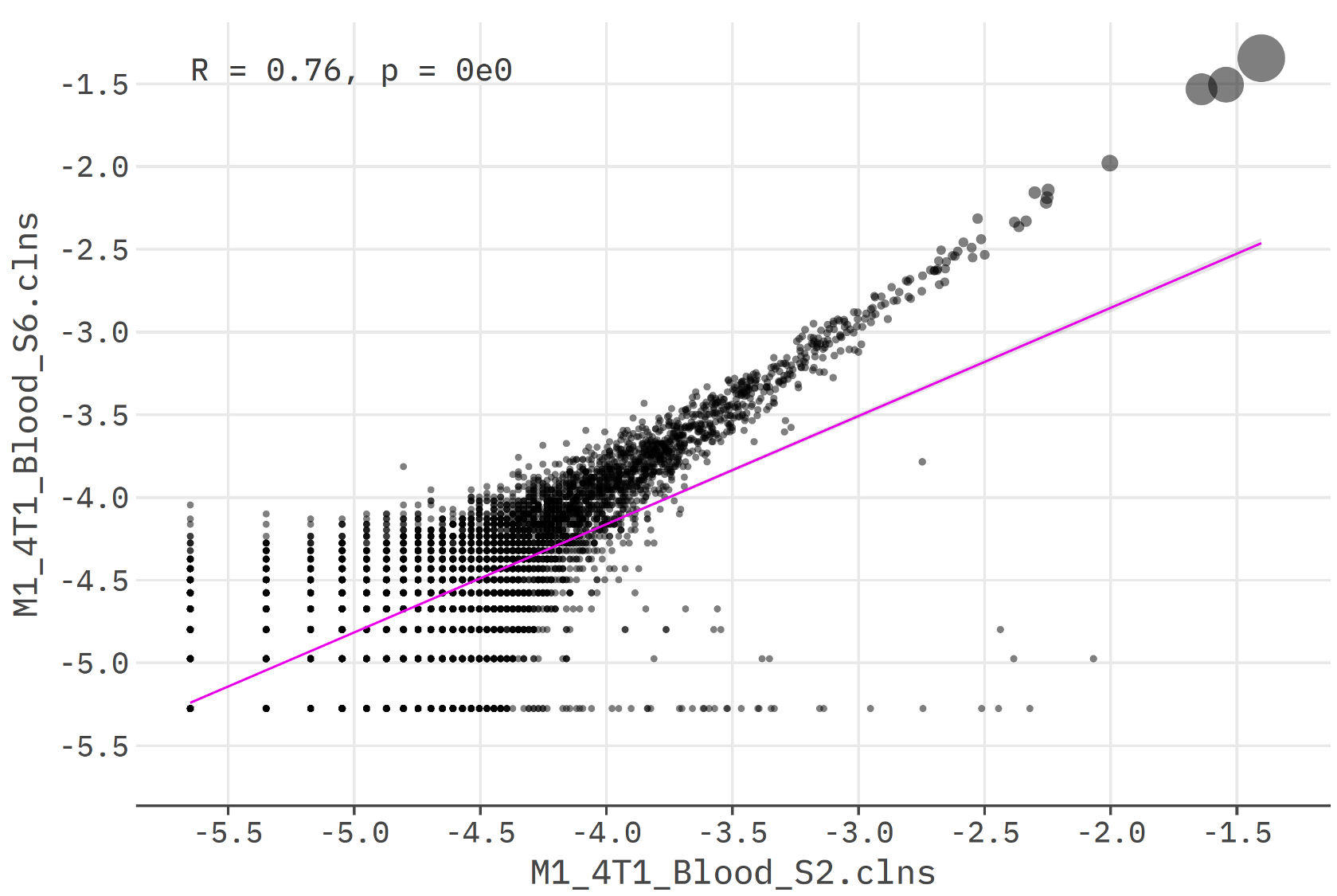
mixcr overlapScatterPlot
mixcr overlapScatterPlot creates a scatter-plot for clone frequencies overlap between two samples.
Command line options
mixcr overlapScatterPlot
--downsampling (<type>|none)
[--chains <chain>[,<chain>...]]...
[--only-productive]
[--criteria <s>]
[--method <method>]
[--no-log]
[--force-overwrite]
[--no-warnings]
[--verbose]
[--help]
cloneset_1.(clns|clna) cloneset_2.(clns|clna) output.(pdf|eps|svg|png|jpeg)
The command takes .clna or .clns file as input and produces one of the following graphical formats depending on the extension of output file: .pdf, .eps, .png, .svg and .jpeg
--downsampling (<type>|none)- Choose downsampling applied to normalize the clonesets. Possible values:
count-[reads|TAG]-[auto|min|fixed][-<number>],top-[reads|TAG]-[<number>],cumtop-[reads|TAG]-[percent],none --chains <chain>[,<chain>...]- Chains to export.
--only-productive- Filter out-of-frame sequences and sequences with stop-codons.
--criteria <s>- Overlap criteria. Defines the rules to treat clones as equal. Default:
CDR3|AA|V|J(For two clones to me equal they must shareCDR3amino acid sequence, V and J genes) --method <method>- Correlation method to use. Possible value: Pearson, Kendall, Spearman. Default: Pearson
--no-log- Do not apply log10 to clonotype frequencies.
-f, --force-overwrite- Force overwrite of output file(s).
-nw, --no-warnings- Suppress all warning messages.
--verbose- Verbose messages.
-h, --help- Show this help message and exit.
Example:
> mixcr overlapScatterPlot \
--downsampling none \
--chains IGH \
results/M1_4T1_Blood_S2.clns results/M1_4T1_Blood_S6.clns \
M1_4T1_Blood.overlap.pdf